Arbovirusbd Database -How to Use
The main purpose of the Arbovirusbd database is to store arbovirus genomic sequences and to increase this data, with additional information obtained from clinical, epidemiological and geographic studies. It was developed through a partnership between Gonçalo Moniz Research Center / Oswaldo Cruz Foundation (Brazil), Bahia State University (Brazil), Rega Institute at Katholieke Universiteit Leuven (Belgium) and University of KwaZulu-Natal (South Africa).
The Database Interface
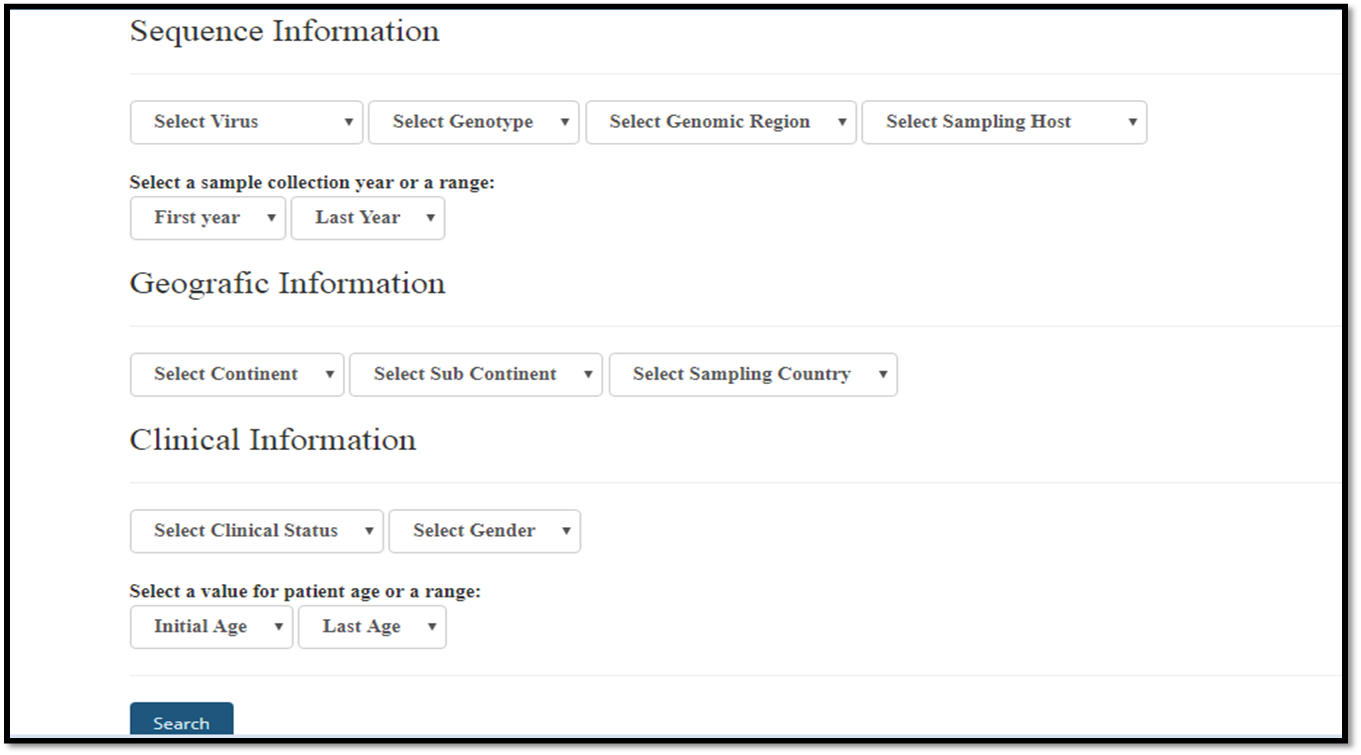
The Arbovirusbd database home page displays an interface that contains multiple fields for refining database queries.
One or more fields can be selected. The user also may select multiple choices to make the search more efficient, to do that hold down the ctrl key.
Start the search by selecting one or more of the types of arboviruses available. if you select multiple virus types simultaneously, you will not see options for the fields: genotypes, genomic regions and clinical status.
The remaining unused fields will be ignored when searching the database, but their values will be presented in the final result.
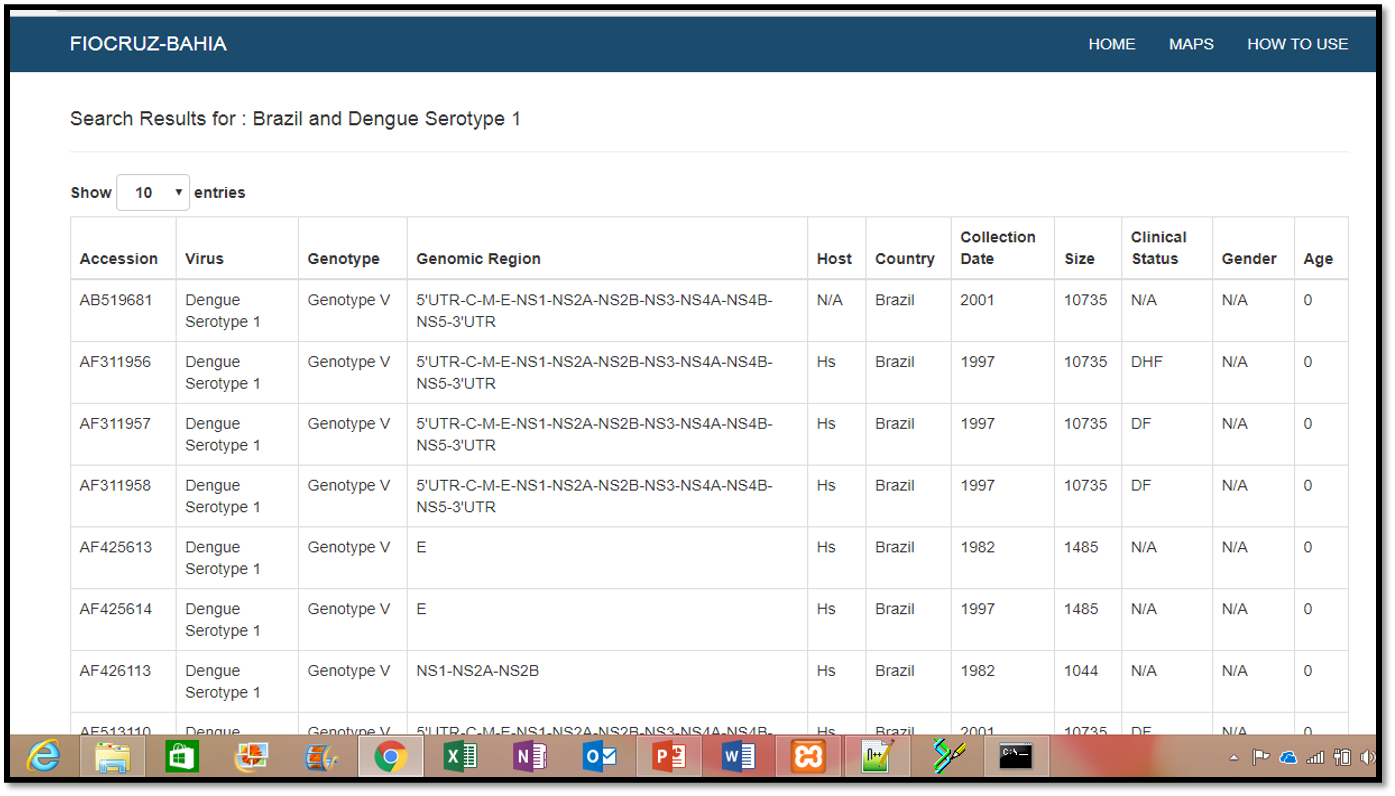
Example
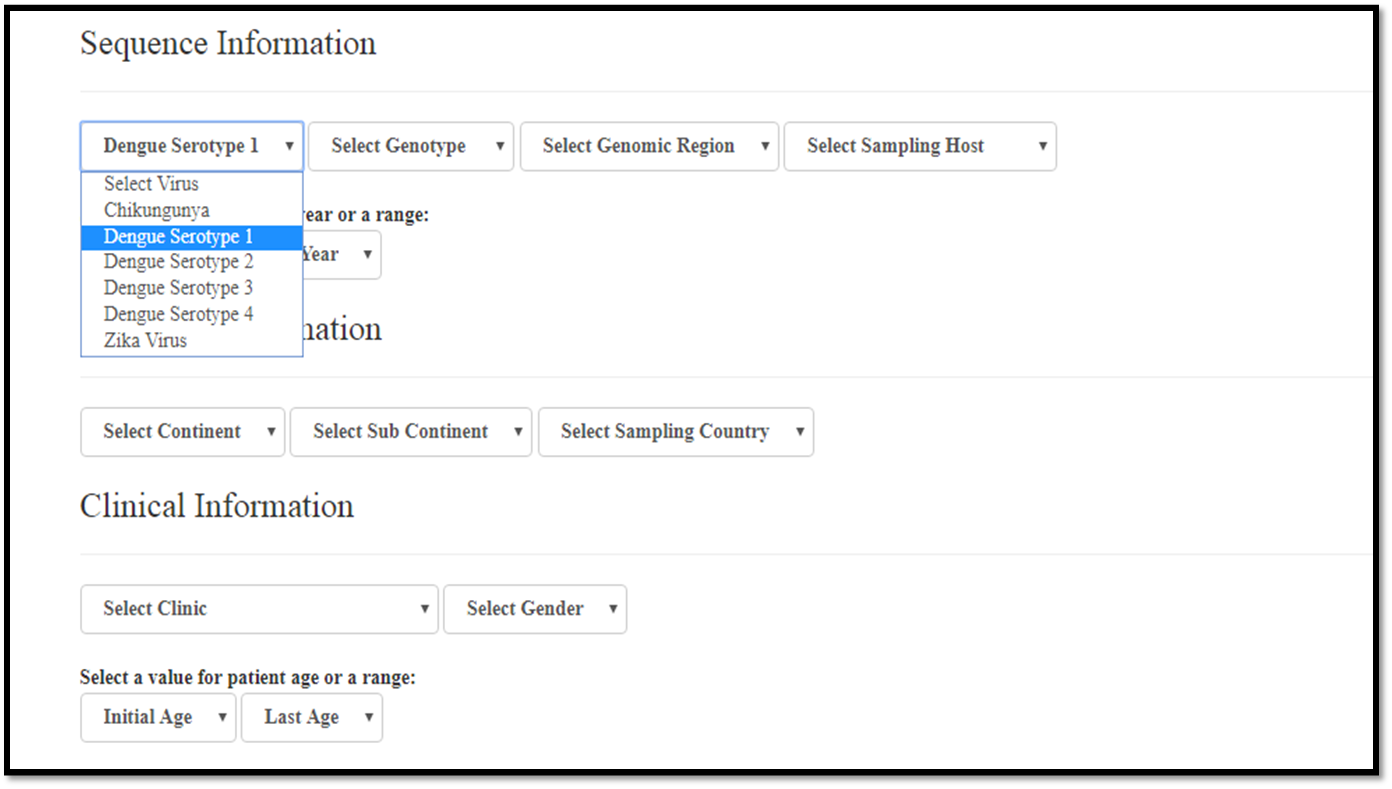
Selecting a virus
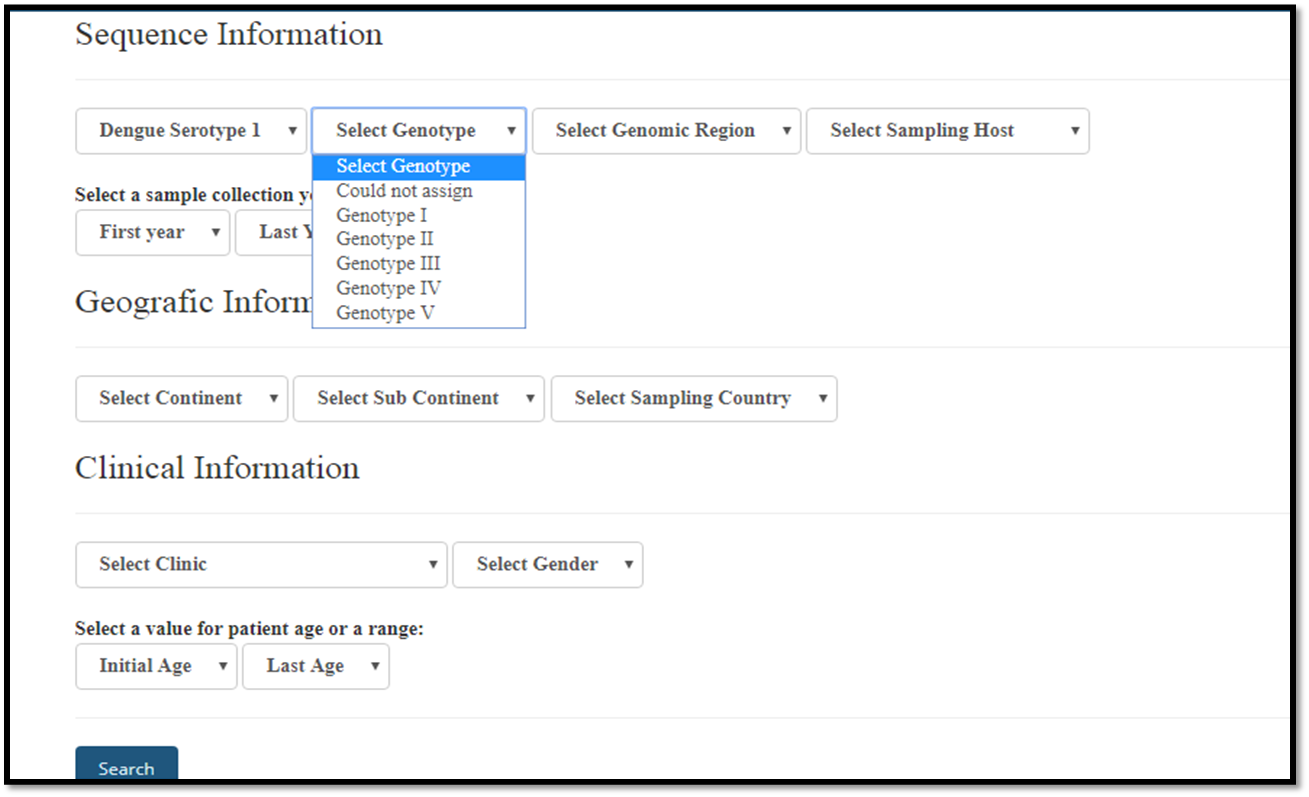
Selecting a genotype
Selecting a genomic region
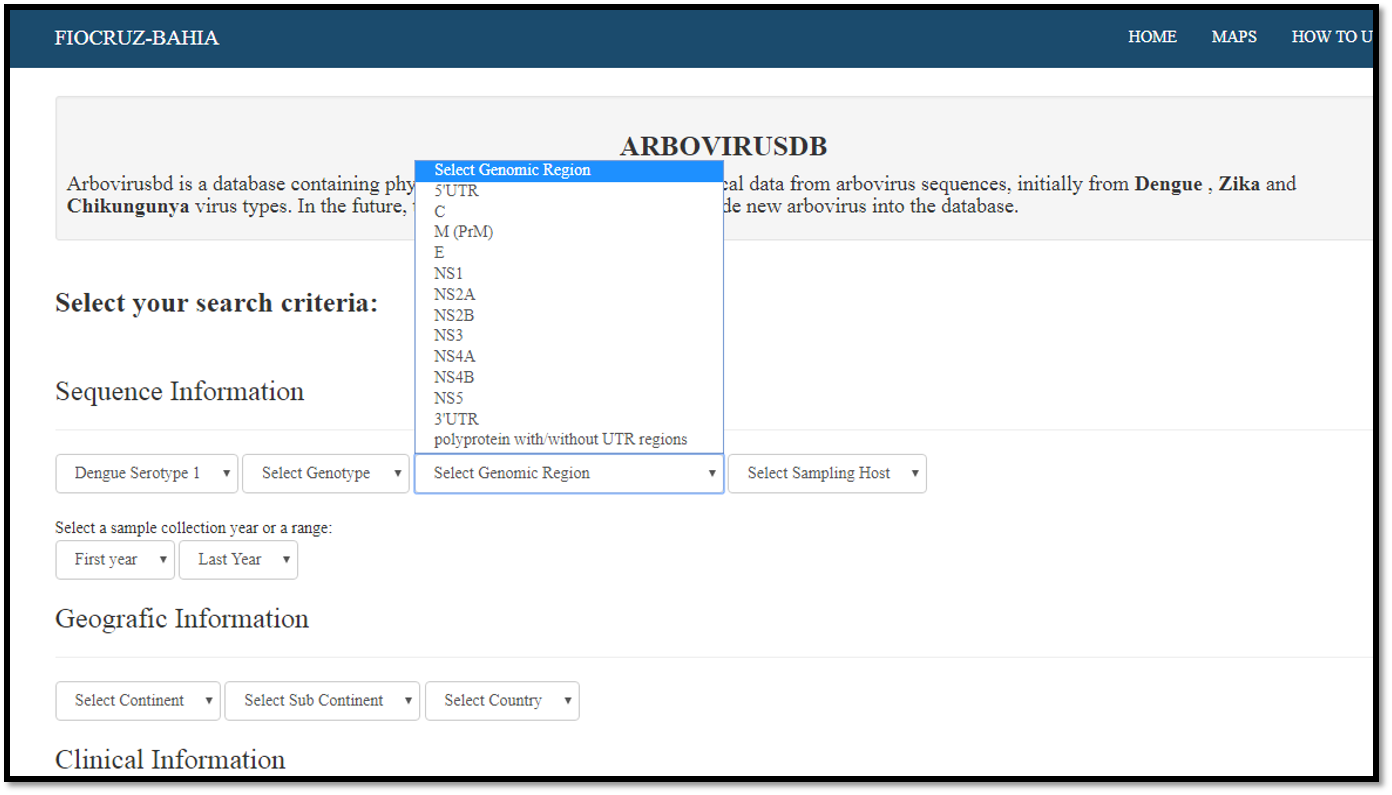
Database Fields Definitions
Virus: initially it is possible to choose between the following arboviruses: ZIKV, CHIKV, DENV-1, DENV-2, DENV-3, DENV-4.
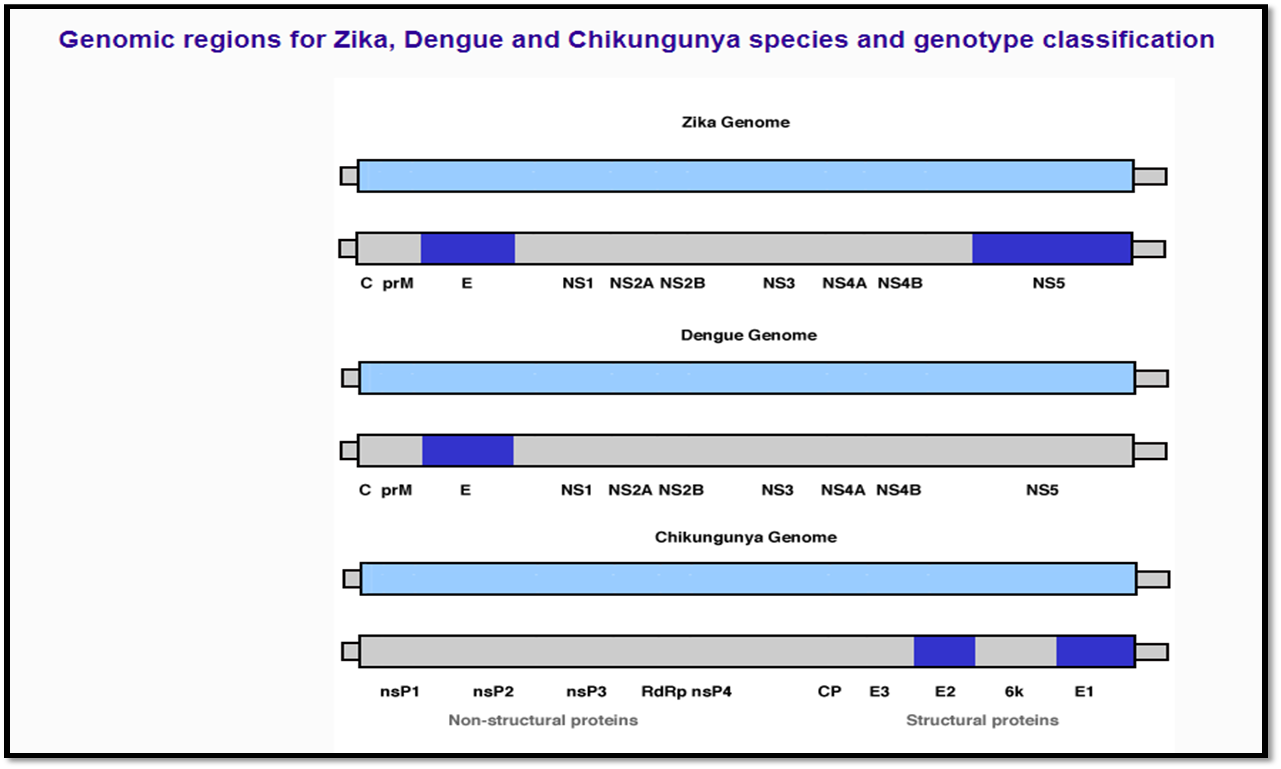
Genotype: shows all genotypes for each selected virus, obtained with the Dengue, Zika & Chikungunya Viruses Typing Tool Version 1.0 .
Genomic Region: shows all available genomic regions in the database for each virus type. Following the nomenclature presented in the Dengue, Zika & Chikungunya Viruses Typing Tool.
Sampling Host: in this field it is possible to select the host from the sequence, when this information is available. For exemple: homo sapiens, mosquito varieties etc.
Sampling Collection Year: the collection year of the sequence or zero when it is not available
Continent: represents the registered continents in the database.
SubContinent: represents the registered subcontinents in the database.
Sampling Country: represents the sampling country registered in the database.
Clinical Status: shows clinical profile values of the host patient sequence or none in other cases.
Gender: this field shows the values M for male, F for female, if the host is homo sapiens and none in other cases
Age: shows the registered age of the patient, if the host is homo sapiens or 0 in other cases.
Data Presentation
After choosing the search criteria, click the search buttom and the results will be shown in table format
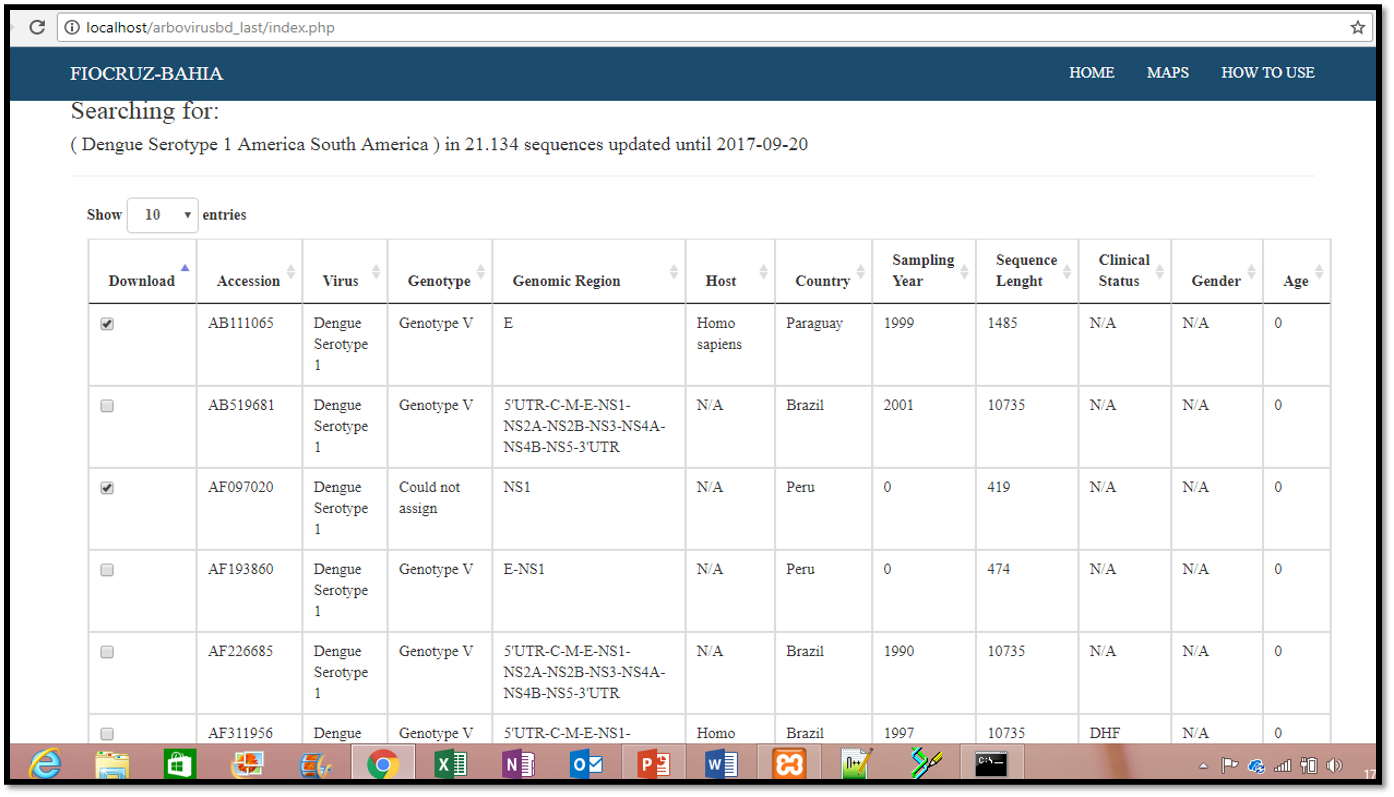
Donwloads
Excel: The search result will be available in standard spreadsheet format, in this tool it is possible perform other file changes.
Fasta: The sequences in FASTA format will begin with a single-line description, followed by lines of sequence data. The first line contains the character '>' and the accession genbank locator, it is possible to add optionally the year and sampling country of each sequence.
Sequences Global Distribution Maps
In this page Clicking on the respective button, it is possible to choose geographic information for each virus.
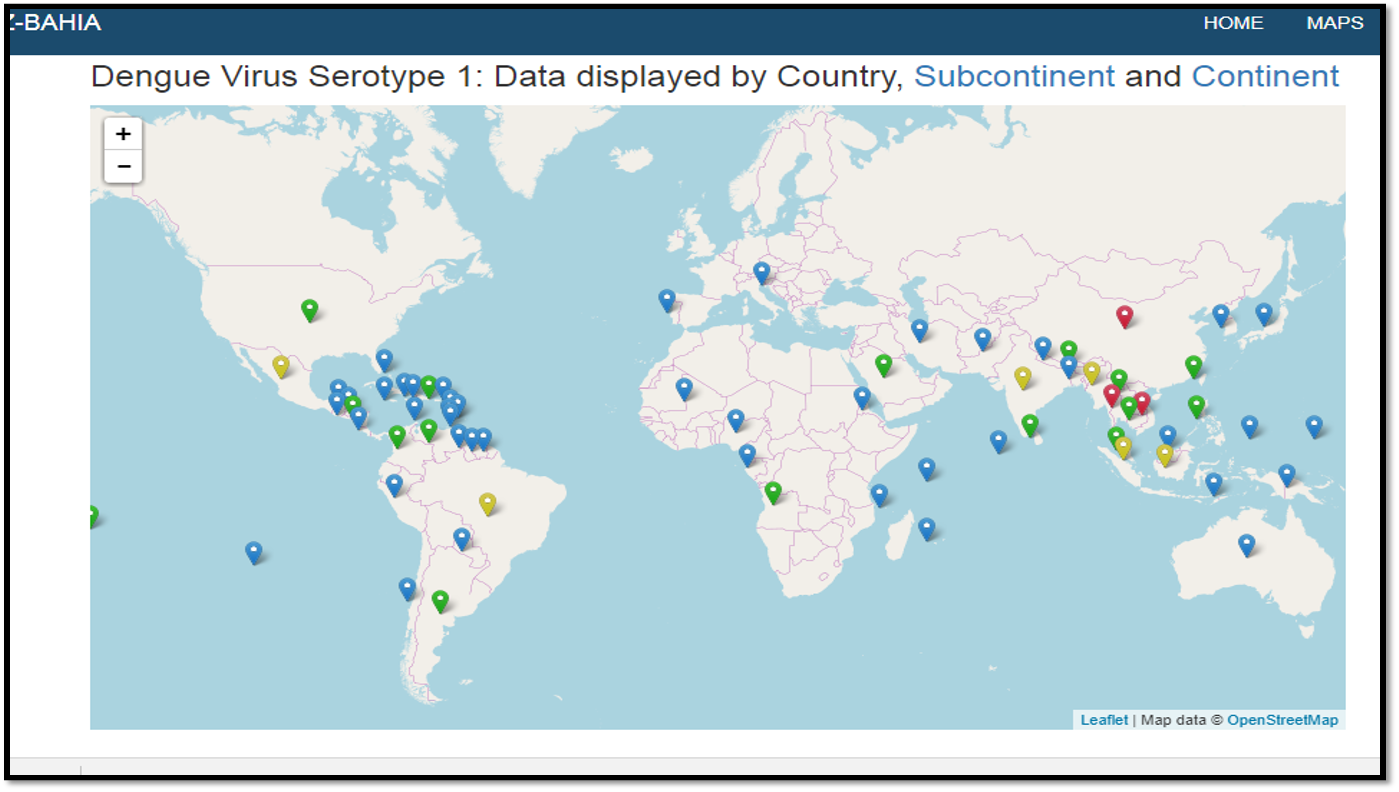
Shows the geographic distribution of each virus and an overview for all viruses and for Dengue. To access the information, click on the pin
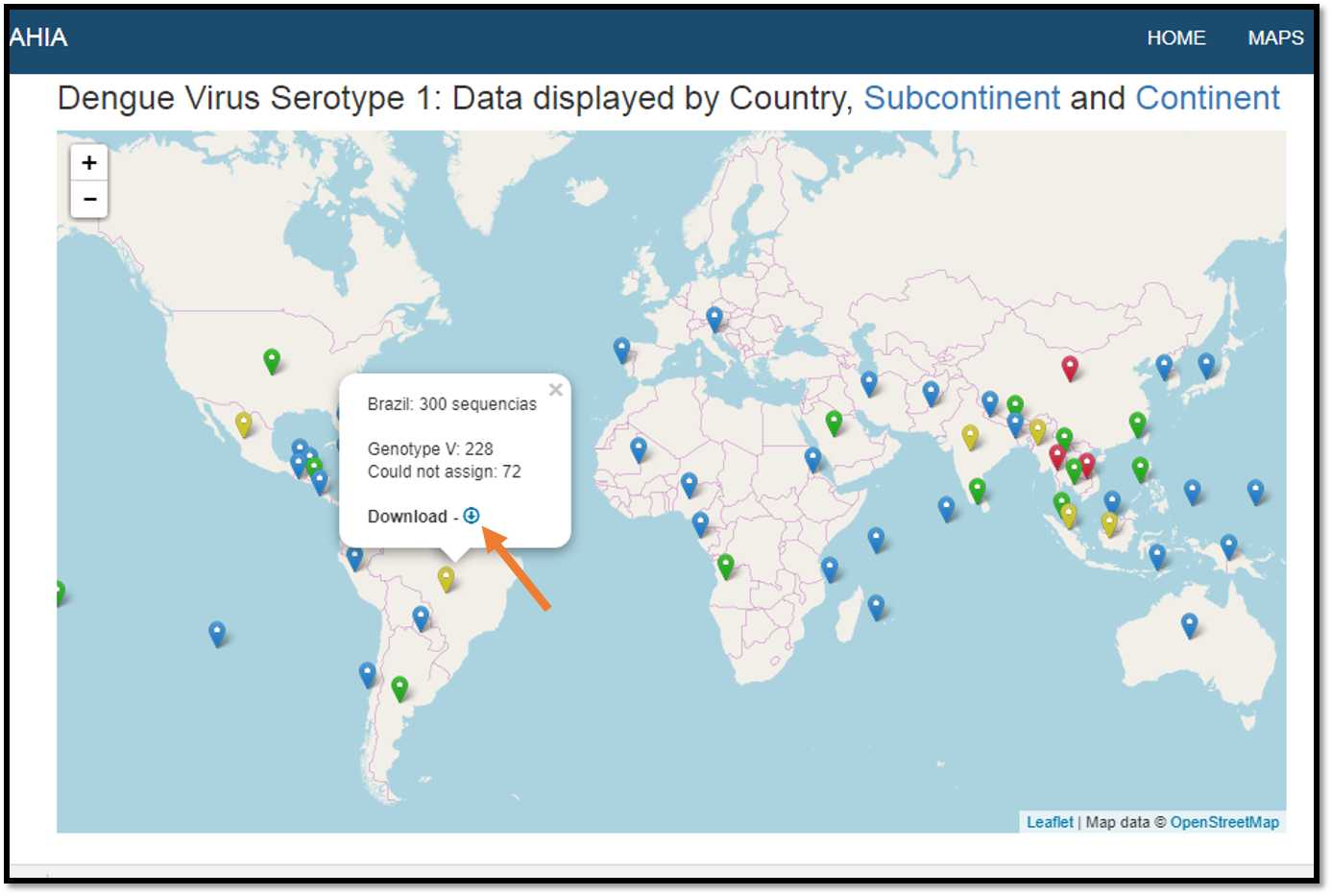
The maps that contain a download symbol in the popup, it is possible to download the data directly from the map